Macromolecular docking is the computational modelling of the quaternary structure of complexes formed by two or more interacting biological macromolecules...
25 KB (3,227 words) - 14:33, 9 October 2024
Macromolecular docking Molecular mechanics Protein structure Protein design Software for molecular mechanics modeling List of protein-ligand docking software...
34 KB (4,011 words) - 19:23, 9 May 2025
The phenomenon of macromolecular crowding alters the properties of molecules in a solution when high concentrations of macromolecules such as proteins...
21 KB (2,498 words) - 03:12, 4 May 2025
prediction Protein structure prediction software Gene prediction Macromolecular docking Protein–DNA interaction site predictor Two-hybrid screening FastContact...
25 KB (2,915 words) - 08:44, 9 May 2024
In molecular biology, the term macromolecular assembly (MA) refers to massive chemical structures such as viruses and non-biologic nanoparticles, cellular...
23 KB (2,293 words) - 16:09, 3 November 2024
Rosetta@home (section RosettaDock)
prediction and protein–protein docking prediction, respectively. Rosetta consistently ranks among the foremost docking predictors, and is one of the best...
86 KB (8,602 words) - 13:31, 28 May 2025
The Cluster of Excellence Frankfurt "Macromolecular Complexes" (CEF) was established in 2006 by Goethe University Frankfurt together with the Max Planck...
118 KB (15,449 words) - 16:35, 18 July 2024
CHARMM (redirect from Chemistry at Harvard Macromolecular Mechanics)
Chemistry at Harvard Macromolecular Mechanics (CHARMM) is the name of a widely used set of force fields for molecular dynamics, and the name for the molecular...
17 KB (2,102 words) - 02:29, 9 March 2025
Structural bioinformatics (section Molecular docking)
such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs...
35 KB (3,532 words) - 23:28, 22 May 2024
necessarily referring to quantum mechanics. Examples are molecular docking, protein-protein docking, drug design, combinatorial chemistry. The fitting of shape...
8 KB (962 words) - 00:43, 5 September 2024
software for computational chemistry, bioinformatics, cheminformatics, docking, pharmacophore searching and molecular simulation. The company's main customer...
3 KB (260 words) - 13:29, 3 December 2024
for high-throughput virtual screening in the cloud. Drug design Macromolecular docking Molecular mechanics Molecular modelling Protein structure Protein...
8 KB (757 words) - 18:15, 23 May 2025
structure prediction, protein function prediction, prediction of macromolecular docking and interactions, network modelling for systems biology and logic-based...
17 KB (1,435 words) - 13:15, 12 December 2023
peptide prediction, homology modeling and loop simulations, flexible macromolecular docking and energy refinement. However the complexity of problems related...
4 KB (454 words) - 18:34, 10 March 2025
and opened in a molecular docking software. There are many programs that can facilitate molecular docking such as AutoDock, DOCK, FlexX, HYDRO, LIGPLOT,...
22 KB (2,439 words) - 21:35, 27 May 2025
Biological data visualization (section Macromolecular)
Computational docking plays a vital role in structural biology, with software providing a user-friendly web platform for modeling various macromolecular interactions...
62 KB (6,687 words) - 00:42, 24 May 2025
techniques include molecular docking, structure-based pharmacophore prediction, and molecular dynamics simulations. Molecular docking is the most used structure-based...
34 KB (4,015 words) - 08:55, 29 May 2025
object-oriented programming library for manipulating and analyzing macromolecular structures, protein complexes and molecular dynamics trajectories A...
2 KB (145 words) - 12:39, 3 February 2024
design. In particular he is a developer of AutoDock, the most widely-used program used for molecular docking. His main research focus areas are HIV drug...
12 KB (1,075 words) - 15:49, 24 May 2025
investigates protein structure, assembly, and interactions within a larger macromolecular assembly. It was originally coined in reference to the use of limited...
13 KB (1,482 words) - 05:55, 11 April 2025
Lossinsky, A. S.; Shivers, R. R. (2004). "Structural pathways for macromolecular and cellular transport across the blood-brain barrier during inflammatory...
26 KB (2,665 words) - 17:20, 25 May 2025
synchrotrons. Since then, cryo-electron microscopy (cryo-EM) of large macromolecular assemblies has been developed. Cryo-EM uses protein samples that are...
105 KB (10,957 words) - 13:53, 25 May 2025
interactions can be modulated using stimuli like ionic strength of the buffer, macromolecular crowding, temperature, pH and electric field. This can lead to reversible...
22 KB (2,637 words) - 06:01, 3 April 2025
sparsely-populated states of macromolecules, which are important in macromolecular interactions but invisible to conventional structural and biophysical...
30 KB (2,752 words) - 23:46, 30 March 2025
for rapid detection and quantitative characterization of reversible macromolecular hetero-associations in solution". Analytical Biochemistry. 346 (1):...
36 KB (4,515 words) - 02:27, 3 December 2024
Peraro M (2014). "New Strategies for Integrative Dynamic Modeling of Macromolecular Assembly". In Karabencheva-Christova T (ed.). Biomolecular Modelling...
133 KB (8,452 words) - 20:08, 29 May 2025
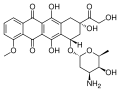
Doxorubicin interacts with DNA by intercalation and inhibition of macromolecular biosynthesis. This inhibits the progression of topoisomerase II, an...
50 KB (4,607 words) - 05:46, 28 May 2025
Motoike H, D'Armiento J, Marks AR, Kass RS (Jan 2002). "Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1...
10 KB (1,213 words) - 02:51, 19 December 2023
Richardson DC (January 2010). "MolProbity: all-atom structure validation for macromolecular crystallography". Acta Crystallographica. Section D, Biological Crystallography...
74 KB (9,238 words) - 21:02, 23 May 2025
Grubmüller, H. (1995). "Predicting slow structural transitions in macromolecular systems: Conformational flooding". Phys. Rev. E. 52 (3): 2893–2906....
33 KB (3,860 words) - 21:00, 25 May 2025