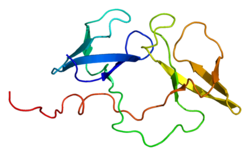
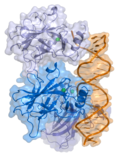
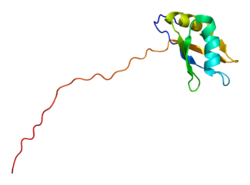
The histone 3′ UTR stem-loop is an RNA element involved in nucleocytoplasmic transport of the histone mRNAs, and in the regulation of stability and of...
3 KB (288 words) - 15:16, 16 January 2020
encodes a protein that binds to the histone 3' UTR stem-loop structure in replication-dependent histone mRNAs. Histone mRNAs do not contain introns or polyadenylation...
11 KB (1,505 words) - 22:19, 15 July 2025
Epigenetics (section Histone modifications)
"Inhibition of G9a Histone Methyltransferase Converts Bone Marrow Mesenchymal Stem Cells to Cardiac Competent Progenitors". Stem Cells International...
159 KB (18,203 words) - 21:52, 9 July 2025
epigenetic modifications. Epigenetic modifications such as DNA methylation and histone modifications alter DNA accessibility and change chromatin structure, thereby...
73 KB (8,905 words) - 13:13, 5 July 2025
polyadenylation factors. Core histone mRNAs have a special stem-loop structure at 3-prime end that is recognized by a stem–loop binding protein and a downstream...
11 KB (1,432 words) - 23:35, 24 July 2024
recruitment of cleavage factors during histone pre-mRNA processing. Duchenne muscular dystrophy Histone 3' UTR stem-loop LSM10 Spycher C, Streit A, Stefanovic...
30 KB (3,630 words) - 03:08, 16 June 2025
preserve neural stem cell pools in the developing brain. Histone acetylation is the addition of acetyl groups on histones by histone acetyltransferases...
69 KB (8,185 words) - 00:47, 16 July 2025
regions of RNA transcripts that fold back on themselves to form short stem-loops (hairpins), whereas siRNAs derive from longer regions of double-stranded...
147 KB (16,635 words) - 21:40, 7 May 2025
Cancer epigenetics (section Histone modification)
called histones to form a structure called a nucleosome. A nucleosome consists of 2 sets of 4 histones: H2A, H2B, H3, and H4. Additionally, histone H1 contributes...
111 KB (12,560 words) - 19:29, 2 July 2025
of approximately 510 nucleotides. The 3' UTR is also predicted to form stem-loops, interior-loops, and bulge-loops, as well as more complex secondary structures...
26 KB (2,537 words) - 03:54, 5 June 2025
Polyadenylation (redirect from Rna 3' polyadenylation signals)
replication-dependent histone mRNAs. These are the only mRNAs in eukaryotes that lack a poly(A) tail, ending instead in a stem-loop structure followed by...
57 KB (7,094 words) - 23:56, 6 August 2024
gene expression. Histone acetylation is performed by histone acetyl transferases (HATs) and histone deacetylation is carried out by histone deacetylases (HDACs)...
97 KB (11,324 words) - 13:10, 15 July 2025
Transcription (biology) (section Enhancers, transcription factors, Mediator complex, and DNA loops in mammalian transcription)
untranslated regions (5'UTR); the sequence after (downstream from) the coding sequence is called the three prime untranslated regions (3'UTR). As opposed to DNA...
58 KB (6,891 words) - 21:22, 4 July 2025
gene can be found on the long (q) arm of the X chromosome at position 27.3, from base pair 146,699,054 to base pair 146,738,156 Almost all cases of fragile...
28 KB (3,412 words) - 11:12, 9 July 2025
: 4.2 The manner in which DNA is stored on the histones, as well as chemical modifications of the histone itself, regulate whether a particular region of...
100 KB (11,213 words) - 16:40, 17 July 2025
embryonic stem cell differentiation. Xist expression is followed by irreversible layers of chromatin modifications that include the loss of the histone (H3K9)...
99 KB (10,625 words) - 08:12, 15 July 2025
human body, multiple stem loops are predicted to occur in the 5' UTR, the coding region of the protein, and in the 3' UTR. The stem loops direct RNA folding...
25 KB (2,172 words) - 03:41, 25 December 2024
P53 (section Stem cells)
Grand RS, Weiss J, Hietter-Pfeiffer E, et al. (June 2023). "Readout of histone methylation by Trim24 locally restricts chromatin opening by p53". Nature...
127 KB (13,874 words) - 22:07, 15 July 2025
the gene itself. Silencers have also been found within the 3 prime untranslated region (3' UTR) of mRNA. Currently, there are two main types of silencers...
27 KB (3,340 words) - 19:51, 25 May 2025
Gene expression (section Enhancers, transcription factors, mediator complex and DNA loops in mammalian transcription)
a 5′ untranslated region (5′UTR), a protein-coding region or open reading frame (ORF), and a 3′ untranslated region (3′UTR). The coding region carries...
104 KB (11,629 words) - 09:41, 17 July 2025
C7orf50 (section Stem Loop Prediction)
5' and 3' UTRs of the mRNA of C7orf50 are predicted to fold into structures such as bulge loops, internal loops, multibranch loops, hairpin loops, and double...
44 KB (3,640 words) - 16:45, 16 July 2025
the SmY ncRNA appears to be involved in mRNA trans-splicing. Y RNAs are stem loops, necessary for DNA replication through interactions with chromatin and...
67 KB (7,183 words) - 05:31, 23 May 2025
based on the extended 5’ and 3’ untranslated region (UTR) in human sequences. Unconserved amino acids, miRNA, stem-loop formations, and RNA binding proteins...
37 KB (3,878 words) - 08:14, 24 June 2025
Possible Stem Loops The 5' UTR has two possible stem loops. These are located from 279-303 and 342-372. In the 3' UTR, there is a possible stem loop located...
12 KB (1,045 words) - 22:24, 15 July 2025
transcripts from such genes are first processed to form the characteristic stem-loop structure of pre-miRNA in the nucleus, then exported to the cytoplasm...
140 KB (15,415 words) - 03:24, 11 June 2025
S (2007). "Stabilization of SMAR1 mRNA by PGA2 involves a stem loop structure in the 5' UTR". Nucleic Acids Research. 35 (18): 6004–6016. doi:10.1093/nar/gkm649...
7 KB (806 words) - 02:01, 19 February 2025
into the GAIT complex, which binds to stem-loop-containing GAIT elements in the 3’-untranslated region (3’- UTR) of diverse inflammatory mRNAs and suppresses...
16 KB (1,950 words) - 18:43, 16 July 2025
Marzluff WF (Dec 1987). "The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability". Molecular...
21 KB (1,770 words) - 15:42, 17 July 2025
or ~3-60 kDa. Because of their single-stranded nature, aptamers are capable of forming many secondary structures, including pseudoknots, stem loops, and...
83 KB (9,541 words) - 12:07, 12 July 2025