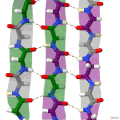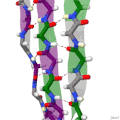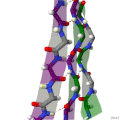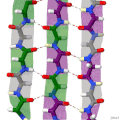
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common secondary structural...
29 KB (3,150 words) - 15:57, 30 April 2025
its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure...
74 KB (9,238 words) - 01:35, 3 April 2025
single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the backbone may interact...
15 KB (1,603 words) - 05:48, 8 February 2025
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed...
38 KB (4,246 words) - 13:59, 17 January 2025
for structure prediction. Detailed list of programs can be found at List of protein secondary structure prediction programs List of protein secondary structure...
6 KB (95 words) - 04:04, 8 May 2025
core of the protein. Secondary structure hierarchically gives way to tertiary structure formation. Once the protein's tertiary structure is formed and...
76 KB (8,708 words) - 09:25, 13 March 2025
A protein contact map represents the distance between all possible amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional...
16 KB (1,979 words) - 07:20, 8 December 2024
Beta sheet (redirect from Greek key (protein structure))
(β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally...
27 KB (3,137 words) - 20:42, 2 March 2025
PSIPRED (section Secondary structure)
PSI-blast based secondary structure PREDiction (PSIPRED) is a method used to investigate protein structure. It uses artificial neural network machine learning...
9 KB (928 words) - 07:59, 12 December 2023
supersecondary structure is a compact three-dimensional protein structure of several adjacent elements of a secondary structure that is smaller than a protein domain...
7 KB (804 words) - 08:29, 8 May 2025
domains of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin...
22 KB (2,602 words) - 15:55, 4 May 2025
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported...
21 KB (2,603 words) - 22:06, 23 November 2024
List of notable protein secondary structure prediction programs List of protein structure prediction software Protein structure prediction...
1 KB (17 words) - 22:00, 5 September 2024
structure of the protein. The tertiary structure is the proteins overall 3D structure which is made of different secondary structures folding together...
40 KB (4,406 words) - 01:13, 15 April 2025
Quinary structure is thus the fifth level of protein complexity, additional to protein primary, secondary, tertiary and quaternary structures. As opposed...
11 KB (1,438 words) - 14:18, 22 August 2024
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure...
21 KB (2,342 words) - 16:59, 19 May 2024
Structural motif (redirect from Secondary structure motif)
chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different...
11 KB (1,202 words) - 06:34, 26 April 2024
310 helix (category Protein structural motifs)
A 310 helix is a type of secondary structure found in proteins and polypeptides. Of the numerous protein secondary structures present, the 310-helix is...
18 KB (1,994 words) - 03:08, 19 August 2023
that share similar amino acid and secondary structure proportions. Each class contains multiple, independent protein superfamilies (i.e. are not necessarily...
9 KB (961 words) - 18:18, 5 April 2025
represent the protein structure allows the solution of many problems including secondary structure prediction, protein-protein interactions, protein-drug interaction...
9 KB (1,423 words) - 11:34, 21 November 2022
Secondary structure prediction is a set of techniques in bioinformatics that aim to predict the secondary structures of proteins and nucleic acid sequences...
879 bytes (133 words) - 11:07, 25 April 2018
responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily...
105 KB (10,957 words) - 19:34, 14 May 2025
biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its...
54 KB (6,283 words) - 06:03, 7 April 2025
active forms, GLP-1 (7–36) amide and GLP-1 (7–37). Active GLP-1 protein secondary structure includes two α-helices from amino acid position 13–20 and 24–35...
29 KB (3,593 words) - 22:16, 9 May 2025
as proteins of known structures, but do not have homologous proteins with known structure. It differs from the homology modeling method of structure prediction...
15 KB (2,013 words) - 22:40, 5 September 2024
three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution...
71 KB (8,443 words) - 19:00, 15 August 2024
Structural alignment (redirect from Protein structure comparison)
more polymer structures based on their shape and three-dimensional conformation. This process is usually applied to protein tertiary structures but can also...
44 KB (5,372 words) - 14:53, 17 January 2025
model error verification List of protein secondary structure prediction programs List of protein structure prediction software Category:Molecular dynamics...
2 KB (174 words) - 17:53, 9 February 2024
Circular dichroism (category Protein structure)
fields. Most notably, far-UV CD is used to investigate the secondary structure of proteins. UV/Vis CD is used to investigate charge-transfer transitions...
52 KB (7,063 words) - 15:30, 3 March 2025
(Homology-derived Secondary Structure of Proteins) is a database that combines structural and sequence information about proteins. This database has...
1,017 bytes (92 words) - 19:25, 16 August 2024