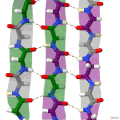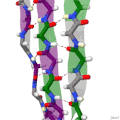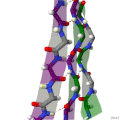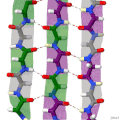
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common secondary structural...
29 KB (3,150 words) - 15:57, 30 April 2025
its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure...
74 KB (9,238 words) - 21:02, 23 May 2025
single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the backbone may interact...
15 KB (1,603 words) - 05:48, 8 February 2025
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed...
38 KB (4,246 words) - 13:59, 17 January 2025
for structure prediction. Detailed list of programs can be found at List of protein secondary structure prediction programs List of protein secondary structure...
6 KB (95 words) - 04:04, 8 May 2025
PSIPRED (section Secondary structure)
PSI-blast based secondary structure PREDiction (PSIPRED) is a method used to investigate protein structure. It uses artificial neural network machine learning...
9 KB (928 words) - 07:59, 12 December 2023
domains of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin...
22 KB (2,602 words) - 15:55, 4 May 2025
core of the protein. Secondary structure hierarchically gives way to tertiary structure formation. Once the protein's tertiary structure is formed and...
76 KB (8,708 words) - 09:25, 13 March 2025
Beta sheet (redirect from Greek key (protein structure))
(β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally...
27 KB (3,137 words) - 20:42, 2 March 2025
A protein contact map represents the distance between all possible amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional...
16 KB (1,979 words) - 07:20, 8 December 2024
urease was in fact a protein. Linus Pauling is credited with the successful prediction of regular protein secondary structures based on hydrogen bonding...
105 KB (10,957 words) - 13:53, 25 May 2025
Secondary structure prediction is a set of techniques in bioinformatics that aim to predict the secondary structures of proteins and nucleic acid sequences...
879 bytes (133 words) - 11:07, 25 April 2018
List of notable protein secondary structure prediction programs List of protein structure prediction software Protein structure prediction...
1 KB (17 words) - 22:00, 5 September 2024
structure of the protein. The tertiary structure is the proteins overall 3D structure which is made of different secondary structures folding together...
40 KB (4,406 words) - 05:02, 27 May 2025
active forms, GLP-1 (7–36) amide and GLP-1 (7–37). Active GLP-1 protein secondary structure includes two α-helices from amino acid position 13–20 and 24–35...
29 KB (3,593 words) - 11:46, 25 May 2025
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported...
21 KB (2,603 words) - 22:06, 23 November 2024
Circular dichroism (category Protein structure)
fields. Most notably, far-UV CD is used to investigate the secondary structure of proteins. UV/Vis CD is used to investigate charge-transfer transitions...
52 KB (7,063 words) - 18:55, 25 May 2025
supersecondary structure is a compact three-dimensional protein structure of several adjacent elements of a secondary structure that is smaller than a protein domain...
7 KB (804 words) - 08:29, 8 May 2025
that share similar amino acid and secondary structure proportions. Each class contains multiple, independent protein superfamilies (i.e. are not necessarily...
9 KB (961 words) - 18:18, 5 April 2025
Structural motif (redirect from Secondary structure motif)
chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different...
11 KB (1,202 words) - 06:34, 26 April 2024
(Homology-derived Secondary Structure of Proteins) is a database that combines structural and sequence information about proteins. This database has...
1,017 bytes (92 words) - 19:25, 16 August 2024
bioinformatics algorithms had to be programmed by hand; for problems such as protein structure prediction, this proved difficult. Machine learning techniques such...
72 KB (8,279 words) - 02:49, 26 May 2025
biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its...
54 KB (6,283 words) - 18:15, 25 May 2025
Polyproline helix (category Protein structural motifs)
A polyproline helix is a type of protein secondary structure which occurs in proteins comprising repeating proline residues. A left-handed polyproline...
10 KB (1,313 words) - 13:58, 24 July 2024
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure...
21 KB (2,342 words) - 16:59, 19 May 2024
STRIDE (algorithm) (redirect from STRIDE (protein))
In protein structure, STRIDE (Structural identification) is an algorithm for the assignment of protein secondary structure elements given the atomic coordinates...
3 KB (381 words) - 03:05, 9 December 2022
model error verification List of protein secondary structure prediction programs List of protein structure prediction software Category:Molecular dynamics...
2 KB (174 words) - 17:53, 9 February 2024
JPred Protein Secondary Structure Prediction Server which provides predictions by the JNet algorithm, one of the most accurate methods for secondary structure...
5 KB (593 words) - 04:24, 1 January 2023
Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as...
18 KB (2,194 words) - 22:57, 24 February 2025
Linus Pauling (section Structure of the atomic nucleus)
worked on the structures of biological molecules, and showed the importance of the alpha helix and beta sheet in protein secondary structure. Pauling's approach...
143 KB (14,181 words) - 05:41, 23 May 2025